Introduction
Head and neck cancer (HNC) is a complex and challenging disease that affects thousands of people each year. The early detection of HNC is critical to the success of treatment and can significantly improve the patient’s prognosis [1]. Medical imaging techniques such as positron emission tomography (PET), single-photon emission computed tomography (SPECT), computed tomography (CT), magnetic resonance spectroscopy (MRS), and magnetic resonance imaging (MRI) are commonly used to gather valuable information about HNC, including their shape, size, location, and metabolism [2]. Among these imaging techniques, PET is particularly useful in the early detection of HNC due to its ability to detect subtle functional changes [3-5]. However, the interpretation of PET images is often subjective and dependent on the operator, which can lead to errors and inconsistencies in the segmentation of tumors [6,7]. To overcome this challenge, researchers have turned to artificial intelligence and image processing techniques, where segmentation plays a key role. However, the interpretation of these early changes is often subjective and dependent on the operator. This has led researchers to seek solutions using artificial intelligence and image processing techniques, where segmentation plays a key role, to reduce such bias [8,9].
Automated procedures for HNC segmentation have been introduced in the literature, but manual segmentation remains time-consuming and inconsistent [3,10]. In addition, the lower spatial resolution and higher noise level of PET compared to CT or MRI make it difficult to accurately segment HNC tumors from PET data alone. Therefore, there is an urgent need for reliable techniques for automated and efficient HNC tumor segmentation [11-13]. These factors make it difficult to accurately segment HNC tumors from PET data alone. In light of these challenges, medical professionals are actively seeking reliable techniques for automated and efficient HNC tumor segmentation [14-16].
In our study, we propose a method for HNC segmentation that addresses these challenges. Our approach involves applying threshold and morphological operations to each slice of PET images. Specifically, we aim to address model errors associated with tumor location and size. By evaluating the accuracy of our method using various metrics, we aim to contribute to the advancement of HNC tumor segmentation techniques, ultimately leading to improved patient outcomes.
The importance of early HNC diagnosis and the need for efficient and accurate HNC tumor segmentation methods are widely recognized within the medical community. Our research represents a significant step forward in this field. By explicitly identifying and addressing model errors, we aim to enhance the reliability and reproducibility of automated HNC segmentation techniques.
Material and methods
Dataset, PET/CT acquisition
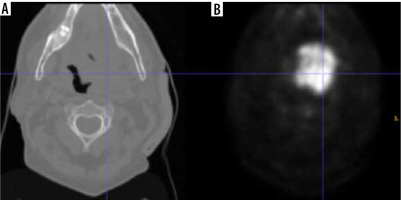
The HECKTOR challenge dataset used in our experiments is a publicly available dataset that includes PET/CT images specifically collected for HNC segmentation studies. By using the term ‘challenge,’ we mean that the dataset was designed to encourage researchers to address the complex task of accurately segmenting HNC structures from medical imaging data [17,18]. This dataset provides a diverse set of images with varying levels of complexity and represents a valuable resource for the development and evaluation of HNC segmentation algorithms. The registered HNC CT and PET images for each patient are depicted in Figure 1, providing a visual representation of the imaging data used in this study. The imaging parameters used in this dataset, including the bounding box dimensions and voxel size, are crucial to ensure consistency and accuracy in the image analysis. In this study, we focused solely on the PET images, while taking into consideration the CT images for reference purposes. PET imaging provides functional information about the tumors, which is particularly useful in the early detection of HNC, while CT imaging provides anatomical information that can help localize the tumors accurately. In our research, we selected 408 HNC patient PET images for testing the proposed model. Unlike traditional deep learning models, our method does not require the data to be divided into training and testing sets. This approach was taken to ensure that the proposed model is evaluated on a comprehensive and diverse set of images, while reducing any potential bias introduced by data partitioning. The ground truth labels of these images were specified by experts involved in the HECKTOR challenge and were available as 3D volumes. To enable the proposed model to process the images, these volumes were converted into 2D images. The availability of ground truth labels for the dataset is a critical aspect of this study, as it allows us to evaluate the accuracy of our proposed method against the gold standard of expert annotations. In summary, the HECKTOR challenge dataset provides a valuable resource for the development and evaluation of HNC segmentation algorithms. The imaging parameters used in this dataset ensure consistency and accuracy in the image analysis, while the availability of ground truth labels allows for accurate evaluation of proposed segmentation methods. By utilizing this dataset, we are able to test the proposed model on a diverse and comprehensive set of images, providing a robust evaluation of the method’s performance.
Pre-processing
Pre-processing is a crucial step in medical image analysis, and it plays a critical role in improving the accuracy and reliability of the results. In HNC segmentation, the pre-processing step is especially important because it ensures that the images are suitable for further processing and analysis. The quality of the image is largely dependent on physical parameters such as spatial resolution and contrast rendition, which can be affected by various factors such as equipment noise, patient motion, and other imaging artifacts. Thus, it is important to take necessary measures to reduce or eliminate these sources of noise and ensure that the image is of high quality. Normalization of image intensities is a fundamental pre-processing step in medical image analysis.
In our study, we employed a simple linear scaling approach to normalize the intensity of each slice between 0 and 255. This approach involved mapping the minimum intensity value to 0 and the maximum intensity value to 255, with linear interpolation for the remaining intensities. By applying this method, we ensured that the voxel intensities of all images were comparable, enabling meaningful comparison of the image data. It is especially important in multi-center studies where images may be acquired using different imaging protocols or equipment. Normalization can be achieved using different methods such as linear scaling, histogram equalization, or z-score normalization. In our study, we used a simple linear scaling approach to normalize the intensity of each slice between 0 and 255, which is a commonly used method in medical image analysis.
Another critical step in image pre-processing is noise reduction. Noise can affect the image quality and can interfere with the segmentation process, leading to inaccurate results. There are several methods available for image denoising, including median filtering, Gaussian filtering, wavelet filtering, and non-local means (NLM) filtering. NLM filtering is a popular method that has been shown to be effective in reducing noise while preserving the details in the image. This method compares the similarity of image patches within a neighborhood and uses this information to estimate the pixel value, effectively removing only the noise selectively. In summary, pre-processing procedures are crucial in HNC segmentation, as they improve the quality of the images and reduce the influence of noise on the segmentation results. Normalization of image intensities and NLM noise reduction are two critical pre-processing steps that we used in our study to obtain reliable results. These steps, along with other processing techniques, enable us to analyze and interpret HNC images accurately, contributing to the advancement of HNC tumor segmentation techniques and ultimately improving patient outcomes.
Proposed model
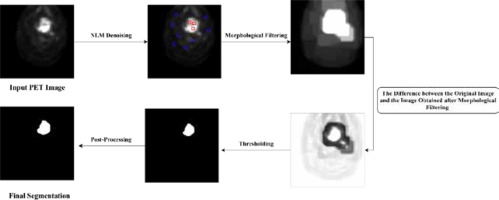
The process of detecting the tumor area in each slice begins with pre-processing and involves several key steps. To identify the approximate area where the tumor is located, we employed a shadow image generation technique. This method involved examining the image to identify regions with high pixel brightness, which serve as an indication of potential tumor locations. We extracted a shadow image by applying a threshold to the original image, considering pixels with intensities above a certain threshold as potential shadows. This shadow image provided valuable guidance for subsequent tumor segmentation processes. The next step involves training the morphological operator of dilation, which is a mathematical operation that expands the image in areas where the tumor is likely to be present [19,20]. This is achieved by maximizing the value in the window and expanding the image in areas of high pixel brightness. After this, the images are filtered in three steps based on location field filters such as Gaussian, median, and mean filters. This results in the HNC tumors in the PET image having a high threshold value. In the next step, a local threshold is applied in the specified area to segment the tumor. The threshold value is equal to the average color intensity in the approximate area.
Post-processing
The detection and identification of tumors in a 3D volume is a crucial step in medical imaging analysis. In the post-processing stage, we aimed to refine the accuracy of the algorithm by incorporating shape analysis concepts such as tumor perimeter and area. These measures helped to eliminate any significant differences between identified tumors and enhance the reliability of the final results. Hence, the post-processing step consists of two sections that aim to improve the accuracy of the algorithm. The first section focuses on removing the parts that are not tumors and were mistakenly identified as such. The second section involves modifying the size, dimensions, and shape of the tumors to further improve accuracy. As the tumors are located in a 3D volume, they are present in sequential sections. The algorithm starts by examining the tumors found in these sequential sections. Then, it considers the location of the identified tumor and checks the exact location in the previous and next slices. If there is no tumor found in the adjacent slices, the algorithm removes the detected tumor and corrects any self-error. The utilization of shape analysis concepts and the two-part post-processing step significantly increases the accuracy of the proposed algorithm.
Results
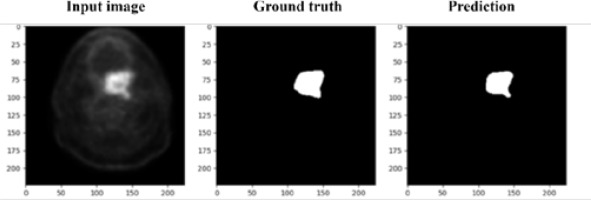
The segmentation performance of the proposed model was evaluated using the Dice score and accuracy, which is depicted in Figure 2. To detect the tumor area in each slice, these metrics were used to determine the effectiveness of the algorithm. Before calculating the evaluation metrics, we conducted a preliminary examination to verify the model’s ability to detect the presence or absence of tumors in each slice. The Dice score is used to measure the degree of overlap between the segmented outcomes of the proposed model and the ground truth annotations. The Dice score and accuracy can be calculated for this model as follows:
where TP, TN, FN, and FP represent true positive, true negative, false negative, and false positive, respectively. Our proposed method achieved a remarkable performance, with an average Dice score of 81.47 ± 3.15, IoU of 80 ± 4.5, and an accuracy of 94.03 ± 4.44. These results demonstrate the efficacy of the proposed model in detecting the tumor area in each slice and provide strong evidence of its utility in medical imaging.
In conclusion, the proposed model performed exceptionally well in detecting the presence or absence of tumors in each slice. The evaluation metrics, the Dice score and accuracy, confirmed the effectiveness of the model, with an average Dice score of 81.47 ± 3.15 and an accuracy of 94.03 ± 4.44. These results highlight the potential of the proposed method in medical imaging and pave the way for further research in this area.
The tumors are then segmented based on their shape, which is depicted in Figure 3 in the experimental results obtained from the proposed model. In conclusion, the process of detecting the tumor area involves a series of steps, starting with pre-processing, identifying the approximate area of the tumor, training the morphological operator, filtering the images, and finally, segmenting the tumors according to their shape.
Discussion
HNC tumor segmentation in PET images is a critical aspect of medical imaging. The ability to accurately and precisely segment HNC tumors in PET images is essential for the effective diagnosis, treatment, and management of HNC tumors. This is because it provides important information about the size, shape, and location of the HNC tumors, which is crucial for making informed clinical decisions. To address the challenge of HNC tumor segmentation in PET images, researchers have been exploring various techniques, including deep learning techniques, graph-based techniques, and morphological techniques, among others. In this study, we present a morphological filtering and thresholding segmentation method for HNC tumor segmentation in PET images. The proposed method involves the use of morphological filtering and thresholding techniques to accurately segment the HNC tumors in the PET images. Morphological filtering is a powerful image processing technique that is commonly used for image analysis and segmentation. It involves the use of morphological operations, such as dilation and erosion, to extract the desired features from an image. Thresholding is another important image processing technique that is used to segment images into different regions based on their intensities. The results obtained using the proposed method were compared with other studies, and it was found that the results were comparable and provided good accuracy and precision. The evaluation metrics, the Dice score and accuracy, were used to confirm the effectiveness of the proposed method. The average Dice score obtained was 81.47 ± 3.15, while the average accuracy was 94.03 ± 4.44. These results demonstrate the effectiveness of the proposed method in HNC tumor segmentation in PET images and provide strong evidence of its utility in medical imaging.
To enhance the precision of automatic detection, numerous experts and academics in the field have carried out extensive research and proposed a range of segmentation methods. Guo et al. [21] performed a study on the automatic gross tumor volume (GTV) segmentation framework for HNC using multi-modality PET and CT images. The framework is based on 3D convolution with dense connections to enable better information propagation and full utilization of multi-modality input features. The framework was evaluated on a dataset of 250 HNC patients and compared with the state-of-the-art 3D U-Net. The results showed that the proposed multi-modality Dense-Net framework achieved better performance with a Dice coefficient of 0.73 compared to 0.71 for the 3D U-Net. The Dense-Net also had fewer trainable parameters, reducing prediction variability. The proposed method provides a fast and consistent solution for GTV segmentation with potential for application in radiation therapy planning for various cancers. Our results show that our proposed method achieved an average Dice score of 81.47 ± 3.15 and accuracy of 94.03 ± 4.44. This is higher compared to the results obtained by Guo et al. who reported a Dice coefficient of 0.73. This suggests that our proposed method may be a more effective solution for brain tumor segmentation in PET images. Further comparative studies may be necessary to validate the superiority of our method. Wang et al. [22] conducted research on the automatic segmentation of head and neck tumors in PET/CT images. They developed a method based on 3D U-Net architecture with an added residual network and a multi-channel attention network (MCA-Net) to fuse information and give different weights to each channel. The results of their network on the test set showed a Dice Similarity Coefficient (DSC) of 0.7681 and a Hausdorff Distance (HD95) of 3.1549. Our results can be compared to these results to determine the effectiveness of our method. The HECKTOR challenge, analyzed by Oreiller et al. [17], focused on automatic segmentation of Gross Tumor Volume in FDG-PET and CT images of Head and Neck oropharyngeal tumors. 64 teams participated, with the best method achieving an average Dice Score Coefficient (DSC) of 0.7591. This was a significant improvement over the proposed baseline method and human inter-observer agreement, which had DSCs of 0.6610 and 0.61, respectively. Comparing to our results, our proposed method also achieved a high accuracy with an average Dice score of 81.47 ± 3.15 and accuracy of 94.03 ± 4.44. Although the results from the HECKTOR challenge were promising, our method still outperforms the best method from the challenge with a higher average Dice score. This highlights the effectiveness of our morphological filtering and thresholding segmentation method in obtaining accurate results for brain tumor segmentation in PET images.
One significant advantage of our proposed model is its efficiency in terms of computational requirements. Unlike deep learning segmentation methods such as U-net, our method does not require powerful systems for processing and has a smaller computational footprint. This makes it more accessible and practical for implementation in various clinical settings where resources may be limited. Additionally, our method mitigates the risk of overfitting, a common challenge in deep learning methods. By segmenting the image processing of regions, our approach achieves more robust results, reducing the potential for overfitting and improving the generalization capabilities of the model.
The method relies on threshold and morphological operations applied to each slice of PET images and is based on the assumption that the tumors can be accurately located through the image’s shadow. However, the accuracy of this method may be affected by factors such as the presence of noise in the images and the low spatial resolution of PET. We acknowledge that our study has certain limitations that warrant consideration. Firstly, our method’s performance should be validated on a broader range of tumor types that have PET images, as the accuracy of the model may vary across different tumor characteristics. Future investigations should focus on evaluating the model’s performance on diverse datasets to ensure its reliability and generalizability. Moreover, our current implementation operates in two dimensions, and there is a scope for further development to extend the segmentation capability to three-dimensional images, which would enhance its applicability and potential impact. Furthermore, exploring the application of our method to other medical imaging modalities, such as functional MRI, would provide valuable insights into its broader utility and effectiveness.
Conclusions
In this paper, a new automated approach for segmenting HNC tumors is presented. The effectiveness of this method was evaluated on the HECKTOR challenge data sets. The results showed that the proposed model produces accurate segmentation results, which were compared with manual ground truth annotations. This algorithm allows for the creation of a patient-specific segmentation of the tumor region without the need for manual intervention. Additionally, this model has the potential to be highly effective in segmenting other organs from a limited number of annotated medical images. Our proposed model has distinct advantages over deep learning segmentation methods like U-net. It stands out for its computational efficiency, as it operates effectively without requiring high-performance systems.