NEURORADIOLOGY / ORIGINAL PAPER
Machine learning-based classification of multiple sclerosis lesion activity using multi-sequence MRI radiomics: a complete analysis of T1, T2, FLAIR, DWI, and SWI features
1
Department of Medical Physics, School of Medicine, Isfahan University of Medical Sciences, Isfahan, Iran
2
Department of Neurology, School of Medicine, Isfahan University of Medical Sciences, Isfahan, Iran
3
Department of Radiology, School of Medicine, Isfahan University of Medical Sciences, Isfahan, Iran
4
Department of Bioimaging, School of Advanced Technologies in Medicine, Isfahan University of Medical Sciences, Isfahan, Iran
Submission date: 2025-04-14
Final revision date: 2025-06-02
Acceptance date: 2025-06-11
Publication date: 2025-08-10
Corresponding author
Daryoush Shahbazi-Gahrouei
Department of Medical Physics, School of Medicine, Isfahan University of Medical Sciences, Isfahan, Iran
Department of Medical Physics, School of Medicine, Isfahan University of Medical Sciences, Isfahan, Iran
Pol J Radiol, 2025; 90: 394-403
KEYWORDS
TOPICS
ABSTRACT
Purpose:
Differentiating active from non-active multiple sclerosis (MS) lesions is critical for disease management but often relies on gadolinium-enhanced magnetic resonance imaging (MRI), raising concerns about retention risks and costs. This study introduces a contrast-free, multi-sequence MRI approach using radiomics and machine learning to classify MS lesion activity.
Material and methods:
A total of 187 lesions from 31 MS patients (mean age 42.5 ± 11.3 years; 64.5% female) at Amin Hospital (November 2024 – February 2025) were retrospectively analysed using a 1.5 T MRI scanner. Five sequences – T1-weighted (T1W), T2-weighted (T2W), fluid-attenuated inversion recovery (FLAIR), diffusion-weighted imaging (DWI), and susceptibility-weighted imaging (SWI) – were processed to extract 8905 radiomic features, refined to 127 via correlation and recursive feature elimination. XGBoost classified lesions as active or non-active, validated on an internal test set (n = 28 lesions), with performance assessed by area under the receiver operating characteristic curve (AUC-ROC).
Results:
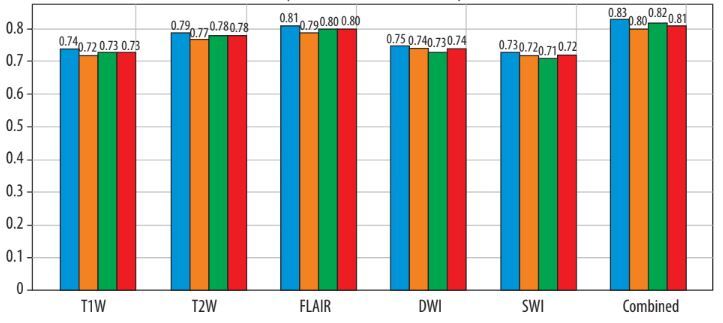
The XGBoost model achieved an AUC-ROC of 0.87 (95% CI: 0.82-0.92), sensitivity of 0.85, and specificity of 0.83, outperforming other classifiers (SVM AUC 0.84). FLAIR (35.4%) and T2W (28.3%) dominated feature contributions, with SWI (12.6%) enhancing accuracy (AUC dropped to 0.84 without SWI). Noise simulation (Gaussian σ = 0.1) confirmed robustness (AUC = 0.86).
Conclusions:
This integration of SWI with conventional sequences in a unified radiomic model offers a promising contrast-free alternative for MS lesion classification, achieving promising accuracy comparable to radiologist performance on an internal test set (n = 28 lesions), pending external validation. External validation is needed to confirm the generalisability, but this approach could reduce gadolinium reliance in clinical practice.
Differentiating active from non-active multiple sclerosis (MS) lesions is critical for disease management but often relies on gadolinium-enhanced magnetic resonance imaging (MRI), raising concerns about retention risks and costs. This study introduces a contrast-free, multi-sequence MRI approach using radiomics and machine learning to classify MS lesion activity.
Material and methods:
A total of 187 lesions from 31 MS patients (mean age 42.5 ± 11.3 years; 64.5% female) at Amin Hospital (November 2024 – February 2025) were retrospectively analysed using a 1.5 T MRI scanner. Five sequences – T1-weighted (T1W), T2-weighted (T2W), fluid-attenuated inversion recovery (FLAIR), diffusion-weighted imaging (DWI), and susceptibility-weighted imaging (SWI) – were processed to extract 8905 radiomic features, refined to 127 via correlation and recursive feature elimination. XGBoost classified lesions as active or non-active, validated on an internal test set (n = 28 lesions), with performance assessed by area under the receiver operating characteristic curve (AUC-ROC).
Results:
The XGBoost model achieved an AUC-ROC of 0.87 (95% CI: 0.82-0.92), sensitivity of 0.85, and specificity of 0.83, outperforming other classifiers (SVM AUC 0.84). FLAIR (35.4%) and T2W (28.3%) dominated feature contributions, with SWI (12.6%) enhancing accuracy (AUC dropped to 0.84 without SWI). Noise simulation (Gaussian σ = 0.1) confirmed robustness (AUC = 0.86).
Conclusions:
This integration of SWI with conventional sequences in a unified radiomic model offers a promising contrast-free alternative for MS lesion classification, achieving promising accuracy comparable to radiologist performance on an internal test set (n = 28 lesions), pending external validation. External validation is needed to confirm the generalisability, but this approach could reduce gadolinium reliance in clinical practice.
REFERENCES (25)
1.
Bandô Y. Mechanism of demyelination and remyelination in multiple sclerosis. Clin Exp Neuroimmunol 2020; 11(S1): 14-21.
2.
Jalilian M, Elhaie M, Sharifi M, Abedi I. Assessment of axonal injury in multiple sclerosis: combined analysis of serum light-chain neurofilaments and diffusion tensor imaging. BMJ Neurol Open 2024; 6: e000788. DOI: 10.1136/bmjno-2024-000788.
3.
Shekari F, Vard A, Adibi I, Danesh-Mobarhan S. Investigating the feasibility of differentiating MS active lesions from inactive ones using texture analysis and machine learning methods in DWI images. Mult Scler Relat Disord 2024; 82: 105363. DOI: 10.1016/j.msard. 2023.105363.
4.
Rostami A, Robatjazi M, Dareyni A, Ghorbani AR, Ganji O, Siyami M, et al. Enhancing classification of active and non-active lesions in multiple sclerosis: machine learning models and feature selection techniques. BMC Med Imaging 2024; 24: 345. doi: 10.1186/s12880-024-01528-6.
5.
Luo W, Huang QX, Huang XW, Matsumoto S, Zeng F, Wang W. Predicting breast cancer in Breast Imaging Reporting and Data System (BI-RADS) ultrasound category 4 or 5 lesions: a nomogram combining radiomics and BI-RADS. Sci Rep 2019; 9: 11921. DOI: 10.1038/s41598-019-48488-4.
6.
Liu Q, Sun D, Li N, Kim JM, Feng D, Huang G, et al. Predicting EGFR mutation subtypes in lung adenocarcinoma using 18F-FDG PET/CT radiomic features. Translat Lung Cancer Res 2020; 9: 549-562.
7.
Rizzo S, Botta F, Raimondi S, Origgi D, Fanciullo C, Morganti AG, et al. Radiomics: the facts and the challenges of image analysis. Eur Radiol Exp 2018; 2: 36. DOI: 10.1186/s41747-018-0068-z.
8.
Ahmadzadeh AM, Lomer NB, Torigian DA. Radiomics and machine learning models for diagnosing microvascular invasion in cholangiocarcinoma: a systematic review and meta-analysis of diagnostic test accuracy studies. Clin Imaging 2025; 121: 110456. DOI: 10.1016/j.clinimag.2025.110456.
9.
Coll L, Pareto D, Carbonell-Mirabent P, Cobo-Calvo Á, Arrambide G, Vidal-Jordana Á, et al. Deciphering multiple sclerosis disability with deep learning attention maps on clinical MRI. Neuroimage Clin 2023; 38: 103376. DOI: 10.1016/j.nicl.2023.103376.
10.
Taloni A, Farrelly FA, Pontillo G, Petsas N, Giannì C, Ruggieri S, et al. Evaluation of disability progression in multiple sclerosis via magnetic-resonance-based deep learning techniques. Int J Mol Sci 2022; 23: 10651. DOI: 10.3390/ijms231810651.
11.
Pilehvari S, Morgan Y, Peng W. An analytical review on the use of artificial intelligence and machine learning in diagnosis, prediction, and risk factor analysis of multiple sclerosis. Mult Scler Relat Disord 2024; 89: 105761. DOI: 10.1016/j.msard.2024.105761.
12.
Elhaie M, Koozari A, Shahbazi-Gahrouei D. Machine learning and neural network approaches for enhanced measuring and prediction of radiation doses. J Radiat Res Appl Sci 2025; 18: 101252. DOI: https://doi.org/10.1016/j.jrra....
13.
Rostami A, Robatjazi M, Dareyni A, Ghorbani AR, Ganji O, Siyami M,et al. Enhancing classification of active and non-active lesions in multiple sclerosis: machine learning models and feature selection techniques. BMC Med Imaging 2024; 24: 345. DOI: 10.1186/s12880-024-01528-6.
14.
Caruana G, Pessini LM, Cannella R, Salvaggio G, de Barros A, Salerno A, et al. Texture analysis in susceptibility-weighted imaging may be useful to differentiate acute from chronic multiple sclerosis lesions. Eur Radiol 2020; 30: 6348-6356.
15.
Dong H, Yang G, Liu F, Mo Y, Guo Y. Automatic brain tumor detection and segmentation using U-net based fully convolutional networks. arXiv:1705.03820 2017. DOI: https://doi.org/10.48550/arXiv....
16.
Bleker J, Roest C, Yakar D, Huisman H, Kwee TC. The effect of image resampling on the performance of radiomics-based artificial intelligence in multicenter prostate MRI. J Magn Res Imaging 2023; 59: 1800-1806.
17.
Jaber HA, Aljobouri HK, Çankaya İ, Koçak OM, Algın O. Preparing fMRI data for postprocessing: conversion modalities, preprocessing pipeline, and parametric and nonparametric approaches. IEEE Access 2019; 7: 122864-122877.
18.
Van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res 2017; 77: e104-e107. DOI: 10.1158/0008-5472.CAN-17-0339.
19.
Ranjbarzadeh R, Kasgari AB, Ghoushchi SJ, Anari S, Naseri M, Bendechache M. Brain tumor segmentation based on deep learning and an attention mechanism using MRI multi-modalities brain images. Sci Rep 2021; 11: 10930. DOI: 10.1038/s41598-021-90428-8.
20.
Moradmand H, Aghamiri SMR, Ghaderi R. Impact of image preprocessing methods on reproducibility of radiomic features in multimodal magnetic resonance imaging in glioblastoma. J Appl Clin Med Phys 2019; 21: 179-190.
21.
Mohammadi-Sadr M, Cheki M, Moslehi M, Zarasvandnia M, Salamat MR. A novel approach based on integrating radiomics, bone morphometry and hounsfield unit-derived from routine chest CT for bone mineral density assessment. Acad Radiol 2025; 32: 2284-2296.
22.
Tripathy RK, Sharma LN, Dandapat S. Diagnostic measure to quantify loss of clinical components in multi-lead electrocardiogram. Healthc Technol Lett 2016; 3: 61-66.
23.
Bonacchi R, Filippi M, Rocca MA. Role of artificial intelligence in MS clinical practice. Neuroimage Clin 2022; 35: 103065. DOI: 10.1016/j.nicl.2022.103065.
24.
Spagnolo F, Depeursinge A, Schädelin S, Akbulut A, Müller H, Barakovic M, et al. How far MS lesion detection and segmentation are integrated into the clinical workflow? A systematic review. Neuroimage Clin 2023; 39: 103491. DOI: 10.1016/j.nicl.2023.103491.
25.
Khajetash B, Talebi A, Bagherpour Z, Abbaspour S, Tavakoli M. Introducing radiomics model to predict active plaque in multiple sclerosis patients using magnetic resonance images. Biomed Phys Eng Express 2023; 9. DOI: 10.1088/2057-1976/ace261.
Share
RELATED ARTICLE
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.